

DNA测序技术在底栖有孔虫监测中的研究进展
收稿日期: 2024-11-25
修回日期: 2025-02-10
网络出版日期: 2025-05-07
基金资助
浙江省海洋水产研究所科技计划项目(HYS-ZX-202410)
Review of DNA Sequencing Technology for Monitoring of Benthic Foraminifera
Received date: 2024-11-25
Revised date: 2025-02-10
Online published: 2025-05-07
Supported by
the Science and Technology Project of Zhejiang Marine Fisheries Research Institute(HYS-ZX-202410)
底栖有孔虫分布广泛、个体小、数量大、物种多样性高、生命周期短,在海洋沉积物中具有良好的保存潜力、并对环境变化具有较高的敏感性,是一种优良的海洋环境质量指示生物。传统的底栖有孔虫监测主要以形态学为主,该方法不仅费时费力,且难以发现一些个体小、丰度低的物种。基于DNA测序的调查方法以其高效、高灵敏度、环境友好等优势,为底栖有孔虫物种鉴定和群落多样性评估提供了新思路。综述了DNA测序技术在底栖有孔虫物种鉴定及分类、群落结构及多样性调查以及大型底栖有孔虫共生体研究等领域的研究进展;指出DNA测序技术在底栖有孔虫监测应用中存在缺乏标准化操作流程、参考数据库不完善、无法绝对定量底栖有孔虫丰度以及高估群落多样性等技术局限性。针对以上局限提出优化建议:制定一套规范统一的操作方案与流程、建立开放共享的底栖有孔虫参考数据库、与荧光定量PCR和eRNA测序技术结合等;未来还应加强基因测序技术的研发和创新,以充分挖掘DNA测序技术在底栖有孔虫监测中的应用潜力。
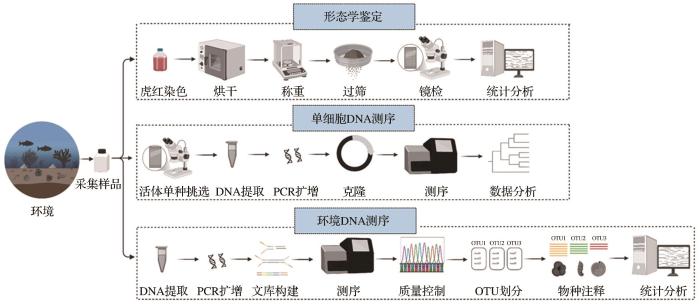
宋静文 , 李铁军 , 郭远明 , 乔玲 . DNA测序技术在底栖有孔虫监测中的研究进展[J]. 地球科学进展, 2025 , 40(3) : 303 -314 . DOI: 10.11867/j.issn.1001-8166.2025.024
Benthic foraminifera are excellent indicators of marine environmental quality due to their wide distribution, small size and large abundance, high species diversity, short life cycle, good preservation potential in marine sediments, and high sensitivity to environmental changes. Traditional monitoring of benthic foraminifera is mainly based on morphology, but this process is time-consuming, labor-intensive, and makes it difficult to detect some species with small individuals and low abundance. The investigation method based on DNA sequencing, with its advantages of high efficiency, high sensitivity, and environmentally friendly, provides a new way to identify benthic foraminifera species and assess community diversity. This paper reviews the progress of DNA sequencing technology in species identification and classification, community structure and diversity investigation, and identification of symbionts in large benthic foraminifera. As there are some technical limitations of DNA sequencing in benthic foraminifera monitoring, such as lack of standardized operation process, imperfection of reference databases, impossibility of absolute quantification of benthic foraminifera abundance, and overestimation of community diversity, optimization can be achieved by formulating a standardized and unified operation process, establishing an open and shared benthic foraminifera reference database, and combining the method with fluorescence quantitative PCR and environmental RNA sequencing technology. In the future, development and innovation of gene sequencing technology should be strengthened to explore the potential of DNA sequencing technology in benthic foraminifera monitoring in a detailed manner.

| 1 | TAPPAN H, LOEBLICH A R. Foraminiferal evolution, diversification, and extinction[J]. Journal of Paleontology, 1988, 62(5): 695-714. |
| 2 | VICKERMAN K. The diversity and ecological significance of protozoa[J]. Biodiversity & Conservation, 1992, 1(4): 334-341. |
| 3 | MALEK M N ABD, FRONTALINI F. Benthic foraminifera as bioindicators of marine pollution: a bibliometric approach to unravel trends, patterns and perspectives[J]. Marine Pollution Bulletin, 2024, 199. DOI: 10.1016/j.marpolbul.2023.115941 . |
| 4 | FRONTALINI F, COCCIONI R. Benthic foraminifera as bioindicators of pollution: a review of Italian research over the last three decades[J]. Revue de Micropaléontologie, 2011, 54(2): 115-127. |
| 5 | LI Tiegang, XIONG Zhifang, JIA Qi. Water exchange between western Pacific warm pool and Indian warm pool and its climatic effects since the late Miocene[J]. Advances in Marine Science, 2020, 38(3): 377-389. |
| 李铁刚, 熊志方, 贾奇. 晚中新世以来印度洋—太平洋暖池水体交换过程及其气候效应[J]. 海洋科学进展, 2020, 38(3): 377-389. | |
| 6 | LI Xiaoyan, SHI Xuefa, CHENG Zhenbo, et al. Distribution of benthic foraminifera in surface sediments of the Laizhou Bay, Bohai Sea and its environmental significance[J]. Acta Micropalaeontologica Sinica, 2010, 27(1): 38-44. |
| 李小艳, 石学法, 程振波, 等. 渤海莱州湾表层沉积物中底栖有孔虫分布特征及其环境意义[J]. 微体古生物学报, 2010, 27(1): 38-44. | |
| 7 | DEBENAY J P, GUILLOU J J, REDOIS F, et al. Distribution trends of foraminiferal assemblages in paralic environments[M]// Environmental micropaleontology. Boston, MA: Springer US, 2000: 39-67. |
| 8 | ZHANG Shuai. The “little giant” in the sea—foraminifera[J]. Knowledge is Power, 2024(6): 14-17. |
| 张帅. 大海里的“小巨人”: 有孔虫[J]. 知识就是力量, 2024(6): 14-17. | |
| 9 | LECROQ B. Molecular assessment of benthic foraminiferal diversity[M]// Approaches to study living foraminifera. Tokyo: Springer Japan, 2013: 91-102. |
| 10 | LI Baohua, KEMAL T E, CHRISTOPH H. Advances in molecular biology of foraminifera[J]. Progress in Natural Science, 2005(5): 534-538. |
| 李保华, Kemal Topac Ertan, Hemleben Christoph. 有孔虫分子生物学研究进展[J]. 自然科学进展, 2005(5): 534-538. | |
| 11 | PAWLOWSKI J. Introduction to the molecular systematics of foraminifera[J]. Micropaleontology, 2000, 46: 1-12. |
| 12 | PAWLOWSKI J, LEJZEROWICZ F, ESLING P. Next-generation environmental diversity surveys of foraminifera: preparing the future[J]. The Biological Bulletin, 2014, 227(2): 93-106. |
| 13 | PAWLOWSKI J, KELLY-QUINN M, ALTERMATT F, et al. The future of biotic indices in the ecogenomic era: integrating (e)DNA metabarcoding in biological assessment of aquatic ecosystems[J]. Science of the Total Environment, 2018, 637: 1 295-1 310. |
| 14 | GOLDSTEIN S T. Foraminifera: a biological overview[M]//Modern foraminifera. Dordrecht: Springer Netherlands, 1999: 37-55. |
| 15 | MEDINGER R, NOLTE V, PANDEY R V, et al. Diversity in a hidden world: potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms[J]. Molecular Ecology, 2010, 19(): 32-40. |
| 16 | ZHANG Kai, LI Baohua, WANG Xiaoyan, et al. Study on the rose Bengal staining for the living benthic foraminifers of the sediments[J]. Acta Micropalaeontologica Sinica, 2020, 37(3): 294-302. |
| 张楷, 李保华, 王晓燕, 等. 沉积物中活体底栖有孔虫的虎红染色方法比较研究[J]. 微体古生物学报, 2020, 37(3): 294-302. | |
| 17 | ZHANG Han, XU Bochao, GUO Xiaoyi, et al. Methods and applications for identifying living benthic foraminifera[J]. Advances in Earth Science, 2021, 36(12): 1 247-1 257. |
| 张涵, 许博超, 郭肖伊, 等. 活体底栖有孔虫鉴别方法及其应用[J]. 地球科学进展, 2021, 36(12): 1 247-1 257. | |
| 18 | MA Weixing, SHA Ou, LIU Yinghong, et al. Study on the supermolecule color reaction of protein and rose Bengal in the presence of emulsifier OP and its analytical applications[J]. Chinese Journal of Analysis Laboratory, 2007, 26(3): 58-62. |
| 马卫兴, 沙鸥, 刘英红, 等. 乳化剂OP存在下蛋白质与虎红的超分子显色反应研究及分析应用[J]. 分析试验室, 2007, 26(3): 58-62. | |
| 19 | SCH?NFELD J, ALVE E, GESLIN E, et al. The FOBIMO (FOraminiferal BIo-MOnitoring) initiative: towards a standardised protocol for soft-bottom benthic foraminiferal monitoring studies[J]. Marine Micropaleontology, 2012, 94: 1-13. |
| 20 | MURRAY J W. Mortality, protoplasm decay rate, and reliability of staining techniques to recognize ‘living’ foraminifera: a review[J]. The Journal of Foraminiferal Research, 2000, 30(1): 66-70. |
| 21 | LANGER M R, LIPPS J H, PILLER W E. Molecular paleobiology of protists: amplification and direct sequencing of foraminiferal DNA[J]. Micropaleontology, 1993, 39(1). DOI:10.2307/1485975 . |
| 22 | BHATT K A, TRIVEDI M H. Molecular studies on foraminifers: past, present, and future[J]. Journal of Foraminiferal Research, 2018, 48(3): 193-209. |
| 23 | PAWLOWSKI J, HOLZMANN M. Diversity and geographic distribution of benthic foraminifera: a molecular perspective[J]. Biodiversity and Conservation, 2008, 17(2): 317-328. |
| 24 | WEBER A A, PAWLOWSKI J. Wide occurrence of SSU rDNA intragenomic polymorphism in foraminifera and its implications for molecular species identification[J]. Protist, 2014, 165(5): 645-661. |
| 25 | QIAO L, FAN S Y, REN C Z, et al. Total and active benthic foraminiferal community and their response to heavy metals revealed by high throughput DNA and RNA sequencing in the Zhejiang coastal waters, East China Sea[J]. Marine Pollution Bulletin, 2022, 184. DOI:10.1016/j.marpolbul.2022.114225 . |
| 26 | QIAO L, CHEN Y, REN C Z, et al. Benthic foraminiferal community structure and its response to environmental factors revealed using high-throughput sequencing in the Zhoushan Fishing Ground, East China Sea[J]. Marine Pollution Bulletin, 2024, 202. DOI:10.1016/j.marpolbul.2024.116385 . |
| 27 | POCHON X, WOOD S A, KEELEY N B, et al. Accurate assessment of the impact of salmon farming on benthic sediment enrichment using foraminiferal metabarcoding[J]. Marine Pollution Bulletin, 2015, 100(1): 370-382. |
| 28 | SHI J F, LEI Y L, LI Q X, et al. Molecular diversity and spatial distribution of benthic foraminifera of the seamounts and adjacent abyssal Plains in the tropical western Pacific Ocean[J]. Marine Micropaleontology, 2020, 156. DOI:10.1016/j.marmicro.2020.101850 . |
| 29 | SCHWEIZER M, PAWLOWSKI J, KOUWENHOVEN T J, et al. Molecular phylogeny of Rotaliida (Foraminifera) based on complete small subunit rDNA sequences[J]. Marine Micropaleontology, 2008, 66(3/4): 233-246. |
| 30 | LI Q X, LEI Y L, LIU J W, et al. Characteristics of foraminiferal communities in the western Clarion-Clipperton Zone revealed by eDNA metabarcoding[J]. Journal of Sea Research, 2022, 189. DOI:10.1016/j.seares.2022.102286 . |
| 31 | LI H T, LEI Y L, LI T G, et al. Next-generation sequencing and metabarcoding to understand the ecology of benthic foraminiferal community in the Bering Sea[J]. Journal of Sea Research, 2023, 191. DOI:10.1016/j.seares.2022.102321 . |
| 32 | LEJZEROWICZ F, GOODAY A J, BARRENECHEA A I, et al. Eukaryotic biodiversity and spatial patterns in the clarion-clipperton zone and other abyssal regions: insights from sediment DNA and RNA metabarcoding[J]. Frontiers in Marine Science, 2021, 8. DOI: 10.3389/fmars.2021.671033 . |
| 33 | MOSS J A, MCCURRY C, SCHWING P, et al. Molecular characterization of benthic foraminifera communities from the Northeastern Gulf of Mexico shelf and slope following the Deepwater Horizon event[J]. Deep Sea Research Part I: Oceanographic Research Papers, 2016, 115: 1-9. |
| 34 | ERTAN K T, HEMLEBEN V, HEMLEBEN C. Molecular evolution of some selected benthic foraminifera as inferred from sequences of the small subunit ribosomal DNA[J]. Marine Micropaleontology, 2004, 53(3/4): 367-388. |
| 35 | Wenlong FA, LI Haotian, LI Qingxia, et al. Preliminary study on the methods of the extraction and pcr amplification of ancient foraminiferal DNA in core sediment from the Yellow Sea[J]. Acta Micropalaeontologica Sinica, 2022, 39(1): 70-84. |
| 法文龙, 李浩天, 李青霞, 等. 黄海沉积物柱状样中有孔虫古DNA的提取和PCR扩增的方法学初探[J]. 微体古生物学报, 2022, 39(1): 70-84. | |
| 36 | LEJZEROWICZ F, ESLING P, PILLET L, et al. High-throughput sequencing and morphology perform equally well for benthic monitoring of marine ecosystems[J]. Scientific Reports, 2015, 5. DOI: 10.1038/srep13932. PMC4564730 . |
| 37 | SHI J F, LEI Y L, LI H T, et al. NGS-metabarcoding revealing novel foraminiferal diversity in the western Pacific Magellan Seamount sediments[J]. Journal of Oceanology and Limnology, 2021, 39(5): 1 718-1 729. |
| 38 | MORARD R, DARLING K F, MAHé F, et al. PFR2: a curated database of planktonic foraminifera 18S ribosomal DNA as a resource for studies of plankton ecology, biogeography and evolution[J]. Molecular Ecology Resources, 2015, 15(6): 1 472-1 485. |
| 39 | BARRENECHEA A I, LEJZEROWICZ F, CORDIER T, et al. Planktonic foraminifera eDNA signature deposited on the seafloor remains preserved after burial in marine sediments[J]. Scientific Reports, 2020, 10(1). DOI: 10.1038/s41598-020-77179-8 . |
| 40 | LEJZEROWICZ F, VOLTSKY I, PAWLOWSKI J. Identifying active foraminifera in the Sea of Japan using metatranscriptomic approach[J]. Deep Sea Research Part II: Topical Studies in Oceanography, 2013, 86: 214-220. |
| 41 | PAWLOWSKI J, ESLING P, LEJZEROWICZ F, et al. Environmental monitoring through protist next-generation sequencing metabarcoding: assessing the impact of fish farming on benthic foraminifera communities[J]. Molecular Ecology Resources, 2014, 14(6): 1 129-1 140. |
| 42 | GRECO M, LEJZEROWICZ F, REO E, et al. Environmental RNA outperforms eDNA metabarcoding in assessing impact of marine pollution: a chromium-spiked mesocosm test[J]. Chemosphere, 2022, 298. DOI: 10.1016/j.chemosphere.2022.134239 . |
| 43 | HOLZMANN M, HABURA A, GILES H, et al. Freshwater foraminiferans revealed by analysis of environmental DNA samples[J]. The Journal of Eukaryotic Microbiology, 2003, 50(2): 135-139. |
| 44 | VOLTSKI I, PAWLOWSKI J. Flexammina islandica gen. nov. sp. nov. and some new phylotypes of monothalamous foraminifera from the coast of Iceland[J]. Zootaxa, 2015, 3 964(2): 245-259. |
| 45 | GOODAY A J, HOLZMANN M, CAULLE C, et al. Giant protists (xenophyophores, Foraminifera) are exceptionally diverse in parts of the abyssal eastern Pacific licensed for polymetallic nodule exploration[J]. Biological Conservation, 2017, 207: 106-116. |
| 46 | VOLTSKI I, WEINER A K M, TSUCHIYA M, et al. Morphological and genetic description of Syringammina limosa sp. nov., the first xenophyophore (Foraminifera) from the deep sea of Okhotsk[J]. Deep Sea Research Part II: Topical Studies in Oceanography, 2018, 154: 32-46. |
| 47 | HOLZMANN M, RIGAUD S, AMINI S, et al. Cyrea szymborska gen. et sp. nov., a new textulariid foraminifer from the Mediterranean sea[J]. Journal of Foraminiferal Research, 2018, 48(2): 156-163. |
| 48 | GOODAY A J, DURDEN J M, HOLZMANN M, et al. Xenophyophores (Rhizaria, Foraminifera), including four new species and two new Genera, from the western Clarion-Clipperton Zone (abyssal equatorial Pacific)[J]. European Journal of Protistology, 2020, 75. DOI: 10.1016/j.ejop.2020.125715 . |
| 49 | AVNAIM-KATAV S, HOLZMANN M, PAWLOWSKI J. Carterina labinea sp. nov.—a new alien foraminifer from the Southeastern Mediterranean shelf[J]. European Journal of Protistology, 2022, 85. DOI: 10.1016/j.ejop.2022.125911 . |
| 50 | GOODAY A J, HOLZMANN M, MAJEWSKI W, et al. New species of Gromia (protista, rhizaria) from south Georgia and the Falkland Islands[J]. Polar Biology, 2022, 45(4): 647-666. |
| 51 | GOODAY A J, HOLZMANN M, SCHWARZGRUBER E, et al. Morphological and molecular diversity of monothalamids (Rhizaria, Foraminifera), including two new species and a new genus, from SW Greenland[J]. European Journal of Protistology, 2022, 86. DOI: 10.1016/j.ejop.2022.125932 . |
| 52 | HOLZMANN M, GOODAY A J, MAJEWSKI W, et al. Molecular and morphological diversity of monothalamous foraminifera from South Georgia and the Falkland Islands: description of four new species[J]. European Journal of Protistology, 2022, 85. DOI: 10.1016/j.ejop.2022.125909 . |
| 53 | KAUSHIK T, DIXIT V, MURUGAN T. Morphology and molecular phylogeny of two new species of Psammophaga (Rhizaria, Foraminifera) from the west coast of India[J]. European Journal of Protistology, 2024, 92. DOI: 10.1016/j.ejop.2023.126035 . |
| 54 | HOLZMANN M. Species concept in foraminifera: ammonia as a case study[J]. Micropaleontology, 2000, 46: 21-37. |
| 55 | WALTON W R, SLOAN B J. The genus Ammonia Bruennich, 1772; its geographic distribution and morphologic variability[J]. The Journal of Foraminiferal Research, 1990, 20(2): 128-156. |
| 56 | TAKATA H, DETTMAN D L, SETO K, et al. Novel habitat preference of ammonia “beccarii” forma 1 in a macrobenthos community on hard substrates in the ohashi river, southwest Japan[J]. The Journal of Foraminiferal Research, 2009, 39(2): 87-96. |
| 57 | HAYWARD B W, HOLZMANN M, TSUCHIYA M. Combined molecular and morphological taxonomy of the beccarii/T3 group of the foraminiferal genus ammonia[J]. Journal of Foraminiferal Research, 2019, 49(4): 367-389. |
| 58 | LEI Y L, LI T G, NIGAM R, et al. Environmental significance of morphological variations in the foraminifer Ammonia aomoriensis (Asano, 1951) and its molecular identification: a study from the Yellow Sea and East China Sea, PR China[J]. Palaeogeography, Palaeoclimatology, Palaeoecology, 2017, 483: 49-57. |
| 59 | KAUSHIK T, MURUGAN T, DAGAR S S. Morphological variation in the porcelaneous benthic foraminifer Quinqueloculina seminula (Linnaeus, 1758): genotypes or morphotypes? A detailed morphotaxonomic, molecular and ecological investigation[J]. Marine Micropaleontology, 2019, 150. DOI: 10.1016/j.marmicro.2019.101748 . |
| 60 | HOHENEGGER J. Growth-invariant meristic characters tools to reveal phylogenetic relationships in Nummulitidae (foraminifera)[J]. Turkish Journal of Earth Sciences, 2011, 20(6): 655-681. |
| 61 | HOLZMANN M, HOHENEGGER J, APOTHéLOZ-PERRET-GENTIL L, et al. Operculina and Neoassilina: a revision of recent nummulitid Genera based on molecular and morphological data reveals a new genus[J]. Journal of Earth Science, 2022, 33(6): 1 411-1 424. |
| 62 | GOODAY A J, HOLZMANN M, GOINEAU A, et al. Xenophyophores (rhizaria, foraminifera) from the eastern clarion-clipperton zone (equatorial Pacific): the genus Psammina [J]. Protist, 2018, 169(6): 926-957. |
| 63 | NGUYEN N L, PAW?OWSKA J, ANGELES I B, et al. Metabarcoding reveals high diversity of benthic foraminifera driven by atlantification of coastal Svalbard[J]. Environmental Science, Biology, 2021. DOI:10.21203/rs.3.rs-1009107/v1 . |
| 64 | FRONTALINI F, GRECO M, di BELLA L, et al. Assessing the effect of mercury pollution on cultured benthic foraminifera community using morphological and eDNA metabarcoding approaches[J]. Marine Pollution Bulletin, 2018, 129(2): 512-524. |
| 65 | WU Yuqi, CHEN Ye, GUO Yuanming, et al. Summary of ecological response of benthic foraminifera to marine environment[J]. Advances in Earth Science, 2024, 39(9): 889-901. |
| 吴玉琦, 陈页, 郭远明, 等. 底栖有孔虫对海洋环境的生态响应概述[J]. 地球科学进展, 2024, 39(9): 889-901. | |
| 66 | PAWLOWSKI J, ESLING P, LEJZEROWICZ F, et al. Benthic monitoring of salmon farms in Norway using foraminiferal metabarcoding[J]. Aquaculture Environment Interactions, 2016, 8: 371-386. |
| 67 | HE X P, SUTHERLAND T F, PAWLOWSKI J, et al. Responses of foraminifera communities to aquaculture-derived organic enrichment as revealed by environmental DNA metabarcoding[J]. Molecular Ecology, 2019, 28(5): 1 138-1 153. |
| 68 | CAVALIERE M, BARRENECHEA A I, MONTRESOR M, et al. Assessing the ecological quality status of the highly polluted Bagnoli area (Tyrrhenian Sea, Italy) using foraminiferal eDNA metabarcoding[J]. Science of the Total Environment, 2021, 790. DOI: 10.1016/j.scitotenv.2021.147871 . |
| 69 | SARASWATI P K. Larger Benthic foraminifera through space and time[M]. Springer Cham: Springer Nature Switzerland AG, 2024: 184. |
| 70 | LEE J J. Fueled by symbiosis, foraminifera have evolved to be giant complex protists[M]// All flesh is grass. Dordrecht: Springer Netherlands, 2010: 427-452. |
| 71 | PRAZERES M, RENEMA W. Evolutionary significance of the microbial assemblages of large benthic Foraminifera[J]. Biological Reviews of the Cambridge Philosophical Society, 2019, 94(3): 828-848. |
| 72 | LANGER M R, LIPPS J H. Phylogenetic incongruence between dinoflagellate endosymbionts (Symbiodinium) and their host foraminifera (Sorites): small-subunit ribosomal RNA gene sequence evidence[J]. Marine Micropaleontology, 1995, 26(1/2/3/4): 179-186. |
| 73 | POCHON X, LaJEUNESSE T C, PAWLOWSKI J. Biogeographic partitioning and host specialization among foraminiferan dinoflagellate symbionts (Symbiodinium; Dinophyta)[J]. Marine Biology, 2004, 146(1): 17-27. |
| 74 | GARCIA-CUETOS L, POCHON X, PAWLOWSKI J. Molecular evidence for host-symbiont specificity in soritid foraminifera[J]. Protist, 2005, 156(4): 399-412. |
| 75 | PAWLOWSKI J, HOLZMANN M, FAHRNI J F, et al. Molecular identification of algal endosymbionts in large miliolid foraminifera: 1. Chlorophytes[J]. The Journal of Eukaryotic Microbiology, 2001, 48(3): 362-367. |
| 76 | PAWLOWSKI J, HOLZMANN M, FAHRNI J F, et al. Molecular identification of algal endosymbionts in large miliolid Foraminifera: 2. Dinofiagellates[J]. The Journal of Eukaryotic Microbiology, 2001, 48(3): 368-373. |
| 77 | HOLZMANN M, BERNEY C, HOHENEGGER J. Molecular identification of diatom endosymbionts in nummulitid Foraminifera[J]. Symbiosis, 2006, 42(2): 93-103. |
| 78 | BRINKMANN I, SCHWEIZER M, SINGER D, et al. Through the eDNA looking glass: responses of fjord benthic foraminiferal communities to contrasting environmental conditions[J]. The Journal of Eukaryotic Microbiology, 2023, 70(4). DOI: 10.1111/jeu.12975 . |
| 79 | SHEN Yangyang, LOU Yanli, LI Haotian, et al. Comparative study on the amplification efficiency of different PCR primers on foraminifera DNA in the sediments of various marine habitats[J]. Acta Micropalaeontologica Sinica, 2020, 37(4): 368-380. |
| 沈阳阳, 类彦立, 李浩天, 等. 不同PCR引物对多种海洋生境沉积物中的有孔虫DNA扩增效能的比较研究[J]. 微体古生物学报, 2020, 37(4): 368-380. | |
| 80 | BORRELLI C, HOU Y B, PAWLOWSKI J W, et al. Assessing SSU rDNA barcodes in foraminifera: a case study using Bolivina quadrata [J]. The Journal of Eukaryotic Microbiology, 2018, 65(2): 220-235. |
| 81 | APOTHéLOZ-PERRET-GENTIL L, CORDONIER A, STRAUB F, et al. Taxonomy-free molecular diatom index for high-throughput eDNA biomonitoring[J]. Molecular Ecology Resources, 2017, 17(6): 1 231-1 242. |
| 82 | ELBRECHT V, LEESE F. Can DNA-based ecosystem assessments quantify species abundance?Testing primer bias and biomass: sequence relationships with an innovative metabarcoding protocol[J]. PLoS ONE, 2015, 10(7). DOI: 10.1371/journal.pone.0130324 . |
| 83 | LOU J, YANG L, WANG H Z, et al. Assessing soil bacterial community and dynamics by integrated high-throughput absolute abundance quantification[J]. PeerJ, 2018, 6. DOI: 10.7717/peerj.4514 . |
| 84 | QIAO L, YU J, LI Y, et al. Amplicon-based illumina sequencing and quantitative PCR reveals nanoplankton diversity and biomass in surface water of Qinhuangdao coastal area, China[J]. Journal of Ocean University of China, 2019, 18(4): 962-976. |
| 85 | ZHU F, MASSANA R, NOT F, et al. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene[J]. FEMS Microbiology Ecology, 2005, 52(1): 79-92. |
| 86 | MURRAY J W. Ecology and applications of benthic foraminifera[M]. Cambridge: Cambridge University Press, 2006. |
| 87 | SEGEV E, SMITH Y, BEN-YEHUDA S. RNA dynamics in aging bacterial spores[J]. Cell, 2012, 148(1/2): 139-149. |
| 88 | LIU T T, YANG H. Comparative analysis of the total and active bacterial communities in the surface sediment of Lake Taihu[J]. FEMS Microbiology Ecology, 2020, 96(5). DOI: 10.1016/S1001-0742(12)60122-3 . |
| 89 | GINER C R, FORN I, ROMAC S, et al. Environmental sequencing provides reasonable estimates of the relative abundance of specific picoeukaryotes[J]. Applied and Environmental Microbiology, 2016, 82(15): 4 757-4 766. |
/
| 〈 |
|
〉 |